10. Epidemics on networks#
We use the Python library “Epidemics on Networks” developed by Kiss, Miller & Simon.
The library must be installed using pip:
pip install EoN
The library documentation is available here.
import EoN
import networkx as nx
import numpy as np
from collections import defaultdict
import pandas as pd
import seaborn as sns
import matplotlib.ticker as ticker
Matplotlib is building the font cache; this may take a moment.
%pylab inline
Populating the interactive namespace from numpy and matplotlib
10.1. Epidemic threshold for homogeneous networks#
We simulate the spread of an SIR on an Erdos-Renyi graph with constant recovery rate.
N = 10000
p = 0.002
G = nx.fast_gnp_random_graph(N, p)
nx.is_connected(G)
True
print(nx.info(G))
Name:
Type: Graph
Number of nodes: 10000
Number of edges: 99584
Average degree: 19.9168
For this network the epidemic threshold can be approximated as \(\lambda_c = \frac{\mu}{\langle k \rangle}\)
mu = 0.2
avg_deg1 = 2 * len(G.edges) / N
lc = mu / avg_deg1
print(lc)
0.01004177377892031
As expected for this network, we have \(\langle k^2 \rangle \sim \langle k \rangle^2 + \langle k \rangle\)
sum_k2 = 0
for i in G.nodes():
k = G.degree(i)
sum_k2 += k * k
avg_k2 = sum_k2 / N
print(avg_k2)
416.0806
avg_deg1**2 + avg_deg1
416.59572224
10.1.1. Simulations of an SIR process#
We simulate 20 realizations of a SIR model for increasing values of \(\lambda\) using the fast_SIR function of EoN
final_size = defaultdict(list)
for lambd in np.geomspace(0.0001, 1.0, 20):
for r in range(0, 20):
t, S, I, R = EoN.fast_SIR(G, lambd, mu, rho=0.05)
final_size[lambd].append(R[-1] / N)
homo_net_size = pd.DataFrame.from_dict(final_size)
homo_net_size
| 0.000100 | 0.000162 | 0.000264 | 0.000428 | 0.000695 | 0.001129 | 0.001833 | 0.002976 | 0.004833 | 0.007848 | 0.012743 | 0.020691 | 0.033598 | 0.054556 | 0.088587 | 0.143845 | 0.233572 | 0.379269 | 0.615848 | 1.000000 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0506 | 0.0514 | 0.0516 | 0.0515 | 0.0524 | 0.0572 | 0.0608 | 0.0700 | 0.0835 | 0.1400 | 0.4589 | 0.7949 | 0.9320 | 0.9829 | 0.9971 | 0.9997 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 1 | 0.0501 | 0.0506 | 0.0514 | 0.0523 | 0.0541 | 0.0548 | 0.0602 | 0.0684 | 0.0938 | 0.1752 | 0.4232 | 0.7858 | 0.9344 | 0.9848 | 0.9981 | 0.9996 | 0.9998 | 1.0 | 1.0 | 1.0 |
| 2 | 0.0506 | 0.0506 | 0.0514 | 0.0521 | 0.0548 | 0.0552 | 0.0614 | 0.0706 | 0.0855 | 0.1488 | 0.3979 | 0.7824 | 0.9336 | 0.9858 | 0.9986 | 0.9999 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 3 | 0.0506 | 0.0507 | 0.0506 | 0.0523 | 0.0537 | 0.0569 | 0.0601 | 0.0684 | 0.0861 | 0.1429 | 0.4395 | 0.7813 | 0.9360 | 0.9852 | 0.9982 | 0.9999 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 4 | 0.0502 | 0.0510 | 0.0518 | 0.0526 | 0.0536 | 0.0557 | 0.0609 | 0.0654 | 0.0922 | 0.1438 | 0.4822 | 0.7811 | 0.9369 | 0.9870 | 0.9981 | 0.9997 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 5 | 0.0507 | 0.0512 | 0.0508 | 0.0516 | 0.0532 | 0.0562 | 0.0593 | 0.0718 | 0.0867 | 0.1415 | 0.4270 | 0.7792 | 0.9354 | 0.9873 | 0.9978 | 0.9999 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 6 | 0.0505 | 0.0507 | 0.0514 | 0.0522 | 0.0538 | 0.0563 | 0.0604 | 0.0672 | 0.0921 | 0.1496 | 0.3933 | 0.7787 | 0.9303 | 0.9867 | 0.9977 | 0.9998 | 0.9999 | 1.0 | 1.0 | 1.0 |
| 7 | 0.0508 | 0.0507 | 0.0513 | 0.0523 | 0.0535 | 0.0561 | 0.0593 | 0.0700 | 0.0932 | 0.1549 | 0.4483 | 0.7800 | 0.9315 | 0.9858 | 0.9982 | 0.9998 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 8 | 0.0506 | 0.0506 | 0.0516 | 0.0516 | 0.0542 | 0.0554 | 0.0613 | 0.0690 | 0.0890 | 0.1517 | 0.4168 | 0.7916 | 0.9378 | 0.9857 | 0.9983 | 1.0000 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 9 | 0.0501 | 0.0504 | 0.0508 | 0.0519 | 0.0537 | 0.0551 | 0.0608 | 0.0674 | 0.1011 | 0.1304 | 0.4228 | 0.7800 | 0.9312 | 0.9867 | 0.9980 | 0.9999 | 0.9999 | 1.0 | 1.0 | 1.0 |
| 10 | 0.0506 | 0.0508 | 0.0511 | 0.0511 | 0.0538 | 0.0568 | 0.0608 | 0.0692 | 0.0893 | 0.1460 | 0.4395 | 0.7839 | 0.9300 | 0.9846 | 0.9976 | 0.9998 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 11 | 0.0510 | 0.0506 | 0.0514 | 0.0515 | 0.0537 | 0.0558 | 0.0593 | 0.0673 | 0.0869 | 0.1531 | 0.4438 | 0.7776 | 0.9413 | 0.9843 | 0.9977 | 0.9996 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 12 | 0.0503 | 0.0503 | 0.0511 | 0.0522 | 0.0541 | 0.0562 | 0.0616 | 0.0685 | 0.0913 | 0.1496 | 0.4670 | 0.7850 | 0.9379 | 0.9842 | 0.9972 | 0.9998 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 13 | 0.0501 | 0.0508 | 0.0511 | 0.0517 | 0.0537 | 0.0544 | 0.0612 | 0.0688 | 0.0858 | 0.1660 | 0.4371 | 0.7954 | 0.9359 | 0.9874 | 0.9981 | 0.9995 | 0.9999 | 1.0 | 1.0 | 1.0 |
| 14 | 0.0503 | 0.0507 | 0.0509 | 0.0523 | 0.0549 | 0.0560 | 0.0587 | 0.0712 | 0.0856 | 0.1583 | 0.4135 | 0.7881 | 0.9309 | 0.9842 | 0.9983 | 0.9996 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 15 | 0.0505 | 0.0505 | 0.0517 | 0.0517 | 0.0544 | 0.0567 | 0.0619 | 0.0701 | 0.0880 | 0.1548 | 0.4127 | 0.7682 | 0.9292 | 0.9872 | 0.9977 | 0.9999 | 0.9999 | 1.0 | 1.0 | 1.0 |
| 16 | 0.0506 | 0.0506 | 0.0509 | 0.0519 | 0.0539 | 0.0569 | 0.0589 | 0.0660 | 0.0886 | 0.1670 | 0.4257 | 0.7781 | 0.9356 | 0.9859 | 0.9981 | 0.9998 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 17 | 0.0500 | 0.0507 | 0.0514 | 0.0525 | 0.0519 | 0.0572 | 0.0613 | 0.0689 | 0.0939 | 0.1540 | 0.4175 | 0.7905 | 0.9329 | 0.9841 | 0.9975 | 0.9995 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 18 | 0.0503 | 0.0506 | 0.0507 | 0.0525 | 0.0525 | 0.0572 | 0.0597 | 0.0689 | 0.0831 | 0.1522 | 0.4397 | 0.7770 | 0.9420 | 0.9841 | 0.9971 | 0.9997 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 19 | 0.0506 | 0.0516 | 0.0508 | 0.0518 | 0.0529 | 0.0564 | 0.0604 | 0.0703 | 0.0853 | 0.1463 | 0.4229 | 0.7737 | 0.9395 | 0.9861 | 0.9986 | 0.9998 | 1.0000 | 1.0 | 1.0 | 1.0 |
plt.figure(figsize=(12, 7))
homo_net_size.boxplot(
positions=np.array(homo_net_size.columns),
widths=np.array(homo_net_size.columns) / 3,
)
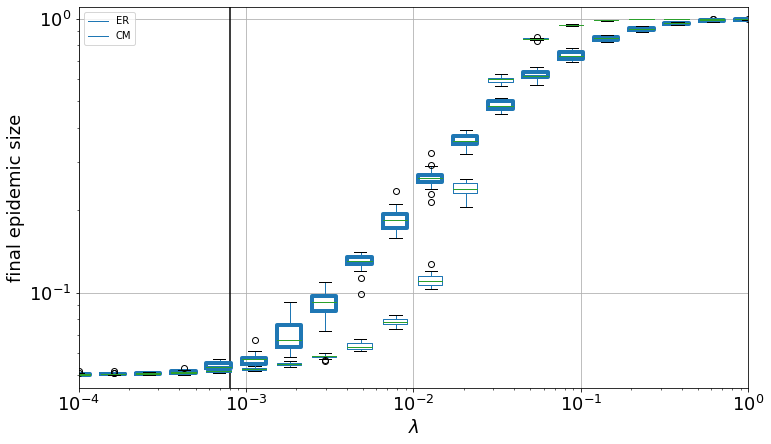
plt.vlines(x=lc, ymin=0.045, ymax=1.1)
plt.xscale("log")
plt.yscale("log")
plt.xlim(0.0001, 1.0)
plt.ylim(0.045, 1.1)
plt.xticks(fontsize=18)
plt.yticks(fontsize=18)
plt.ylabel("final epidemic size", fontsize=18)
plt.xlabel("$\lambda$", fontsize=18)
Text(0.5, 0, '$\\lambda$')
10.2. Epidemic threshold for Barabàsi-Albert model networks#
N = 10000
AB = nx.barabasi_albert_graph(N, 10)
nx.is_connected(AB)
True
print(nx.info(AB))
Name:
Type: Graph
Number of nodes: 10000
Number of edges: 99900
Average degree: 19.9800
sum_k2 = 0
for i in AB.nodes():
k = AB.degree(i)
sum_k2 += k * k
avg_k2 = sum_k2 / N
print(avg_k2)
940.6206
avg_deg = 2 * len(AB.edges) / N
print(avg_deg)
19.98
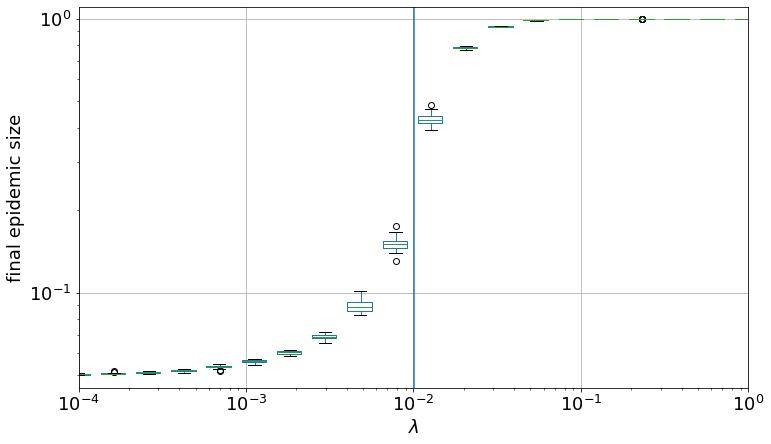
The threshold can be approximated as \(\lambda_c \sim \mu \frac{\langle k \rangle}{\langle k^2 \rangle - \langle k \rangle}\)
lambda_c = mu * avg_deg / (avg_k2 - avg_deg)
print(lambda_c)
0.0043404559824974055
lambda_c / lc
0.43223996856102154
10.2.1. Simulations of an SIR process#
We simulate 20 realizations of a SIR model for increasing values of \(\lambda\)
final_size_AB = defaultdict(list)
for lambd in np.geomspace(0.0001, 1.0, 20):
for r in range(0, 20):
t, S, I, R = EoN.fast_SIR(AB, lambd, mu, rho=0.05)
final_size_AB[lambd].append(R[-1] / N)
sf_net_size = pd.DataFrame.from_dict(final_size_AB)
sf_net_size.tail()
| 0.000100 | 0.000162 | 0.000264 | 0.000428 | 0.000695 | 0.001129 | 0.001833 | 0.002976 | 0.004833 | 0.007848 | 0.012743 | 0.020691 | 0.033598 | 0.054556 | 0.088587 | 0.143845 | 0.233572 | 0.379269 | 0.615848 | 1.000000 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 0.0505 | 0.0507 | 0.0515 | 0.0528 | 0.0533 | 0.0570 | 0.0596 | 0.0824 | 0.1225 | 0.2423 | 0.4803 | 0.7067 | 0.8738 | 0.9585 | 0.9909 | 0.9986 | 0.9999 | 1.0 | 1.0 | 1.0 |
| 16 | 0.0507 | 0.0507 | 0.0514 | 0.0526 | 0.0539 | 0.0585 | 0.0647 | 0.0796 | 0.1165 | 0.2618 | 0.4837 | 0.7169 | 0.8759 | 0.9585 | 0.9923 | 0.9988 | 0.9998 | 1.0 | 1.0 | 1.0 |
| 17 | 0.0507 | 0.0506 | 0.0511 | 0.0519 | 0.0541 | 0.0572 | 0.0654 | 0.0877 | 0.1462 | 0.2727 | 0.4790 | 0.7197 | 0.8774 | 0.9599 | 0.9919 | 0.9987 | 0.9996 | 1.0 | 1.0 | 1.0 |
| 18 | 0.0508 | 0.0505 | 0.0510 | 0.0519 | 0.0539 | 0.0552 | 0.0641 | 0.0813 | 0.1444 | 0.2636 | 0.5012 | 0.7044 | 0.8731 | 0.9540 | 0.9919 | 0.9991 | 1.0000 | 1.0 | 1.0 | 1.0 |
| 19 | 0.0502 | 0.0510 | 0.0508 | 0.0525 | 0.0537 | 0.0555 | 0.0579 | 0.0779 | 0.1541 | 0.2538 | 0.5092 | 0.7004 | 0.8743 | 0.9579 | 0.9923 | 0.9991 | 0.9996 | 1.0 | 1.0 | 1.0 |
plt.figure(figsize=(12, 7))
homo_net_size.boxplot(
positions=np.array(homo_net_size.columns),
widths=np.array(homo_net_size.columns) / 3,
color="r",
)
plt.vlines(x=lambda_c, ymin=0.04, ymax=1.1)
sf_net_size.boxplot(
positions=np.array(sf_net_size.columns), widths=np.array(sf_net_size.columns) / 3
)
plt.yscale("log")
plt.xscale("log")
plt.xlim(0.0001, 1.0)
plt.ylim(0.045, 1.1)
plt.xticks(fontsize=18)
plt.yticks(fontsize=18)
plt.ylabel("final epidemic size", fontsize=18)
plt.xlabel("$\lambda$", fontsize=18)
Text(0.5, 0, '$\\lambda$')
10.3. Epidemic threshold for configuration model networks#
We can use the configuration model to generate a network with a given power-law degree distribution. By setting \(\gamma=2.1\), we expect to see more fluctuations in the degree distribution.
We create a function that samples from a power-law distribution between \(k_{min}\) and \(k_{max}\) with exponent \(\gamma\)
def get_activity(x0, x1, g):
y = random.uniform(0, 1)
e = g + 1.0
a = ((x1**e - x0**e) * y + x0**e) ** (1.0 / e)
return a
N = 10000
kmin = 2
kmax = N - 1
a = []
for i in range(N):
act = get_activity(kmin, kmax, -2.1)
a.append(int(round(act)))
# we need the sum of the degree sequence to be even to properly run the configuration model
if sum(a) % 2 == 0:
G1 = nx.configuration_model(a)
else:
a[-1] += 1
G1 = nx.configuration_model(a)
nx.is_connected(G1)
True
print(nx.info(G1))
Name:
Type: MultiGraph
Number of nodes: 10000
Number of edges: 65624
Average degree: 13.1248
G1 = nx.Graph(G1)
G1.remove_edges_from(nx.selfloop_edges(G1))
print(nx.info(G1))
Name:
Type: Graph
Number of nodes: 10000
Number of edges: 54022
Average degree: 10.8044
nx.is_connected(G1)
True
sum_k2 = 0
for i in G1.nodes():
k = G1.degree(i)
sum_k2 += k * k
avg_k2 = sum_k2 / N
print(avg_k2)
2428.7296
avg_deg = 2 * len(G1.edges) / N
print(avg_deg)
10.8044
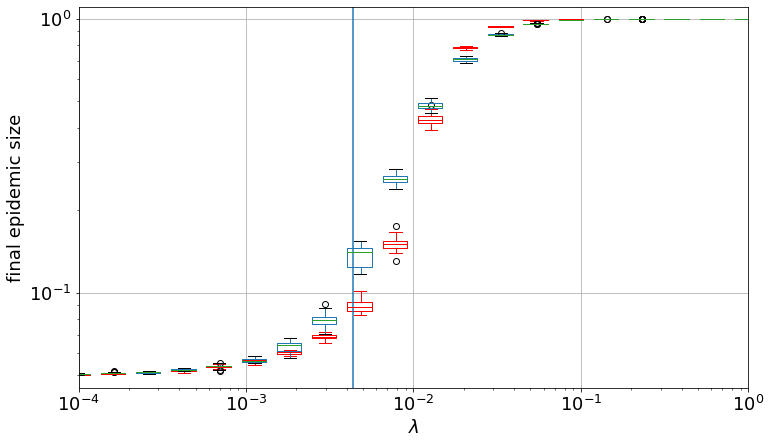
lambda_c1 = mu * avg_deg / (avg_k2 - avg_deg)
print(lambda_c1)
0.000893691831327123
lc / lambda_c1
11.236282381599462
10.3.1. Simulations of an SIR process#
We simulate 100 realizations of a SIR model for increasing values of \(\lambda\)
final_size_conf = defaultdict(list)
for lambd in np.geomspace(0.0001, 1.0, 20):
for r in range(0, 20):
t, S, I, R = EoN.fast_SIR(G1, lambd, mu, rho=0.05)
final_size_conf[lambd].append(R[-1] / N)
conf_net_size = pd.DataFrame.from_dict(final_size_conf)
plt.figure(figsize=(12, 7))
boxprops1 = dict(linestyle="-", linewidth=4, color="b")
homo_net_size.boxplot(
positions=np.array(homo_net_size.columns),
widths=np.array(homo_net_size.columns) / 3,
)
conf_net_size.boxplot(
positions=np.array(conf_net_size.columns),
widths=np.array(conf_net_size.columns) / 3,
boxprops=boxprops1,
)
plt.vlines(x=lambda_c1, ymin=0.04, ymax=1.1)
plt.legend(["ER", "CM"])
plt.yscale("log")
plt.xscale("log")
plt.xlim(0.0001, 1.0)
plt.ylim(0.045, 1.1)
plt.xticks(fontsize=18)
plt.yticks(fontsize=18)
plt.ylabel("final epidemic size", fontsize=18)
plt.xlabel("$\lambda$", fontsize=18)
Text(0.5, 0, '$\\lambda$')